Steven Rokita
Professor
Contact Information
- rokita@jhu.edu
- Curriculum Vitae
- Remsen 124
- by appointment
- 410-516-5793
- Group/Lab Website
Research Interests: Bioorganic chemistry and biochemistry
Education: PhD, Massachusetts Institute of Technology
Steven Rokita received a B.S. (Chemistry) from the University of California at Berkeley and his Ph.D. in Biological Chemistry from MIT under the direction of Christopher Walsh. He joined the laboratory of E. T. Kaiser (Rockefeller University) as a NIH Postdoctoral Fellow in 1983 and then began his independent career in 1985 as a faculty member of the Chemistry Department at what was then called SUNY at Stony Brook. In 1995, he moved as an associate professor to the Department of Chemistry and Biochemistry at the University of Maryland, College Park and most recently (2012) moved as a professor of Chemistry to Johns Hopkins University where he is also director of the Chemistry-Biology Interface Graduate Training Program.
Professor Rokita’s research program is united by a common interest in describing the structure and activity of biological macromolecules through their essential chemical reactivity. The methods of organic synthesis, physical organic chemistry, protein and nucleic acid chemistry, biochemistry and molecular biology are applied to questions on enzyme catalysis and nucleic acid modification. Current projects include enzymatic dehalogenation and reversible covalent chemistry expressed by quinone methide intermediates.
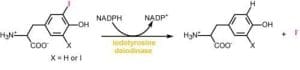
Reductive dehalogenation in nature. Organohalides are distributed widely in the environment and originate from both biological and industrial sources. Under aerobic conditions, these compounds are typically consumed by oxidative and hydrolytic metabolism. However, vertebrates also express the unusual ability to promote reductive dehalogenation of natural iodotyrosine derivatives related to the thyroid hormone thyroxine. Iodide recovery from these compounds is promoted by iodotyrosine deiodinase, an enzyme crucial for human health. This same enzyme has also been discovered in all metazoa sequenced to date as well as some bacteria. The role of reductive dehalogenation in these additional organisms is not yet clear. Investigations are currently designed to identify the origins of this enzyme, characterize its biological role in non-vertebrates and understand its catalytic mechanism that includes a role of its bound flavin. Flavin-dependent reactions are associated with a wide variety of metabolic transformations but its use in reductive dehalogenation is quite rare. Results of this research will help to expand the known repertoire of flavin chemistry and will guide its future use in bioremediation of organohalide pollutants.
Reversible alkylation by quinone methides. Quinone methides (QM) are highly electrophilic and transient intermediates that can form in vivo as byproducts of metabolism. These intermediates are responsible for DNA alkylation observed after exposure to drugs such as tamoxifen, food additives such as butylated hydroxytoluene (BHT), and certain natural products such as mitomycin C. Our laboratory has capitalized on the reversible nature of ortho-QM intermediates in a design for sequence specific alkylation that is compatible with a cellular environment. Internal trapping of the QM is highly effective at suppressing off-target reaction and allows for its transfer only after association with a chosen target. Applicability of this strategy is boosted by the remarkable resistant of QM transfer to high concentrations of thiols like those present in our cells to protect against damage by electrophiles.
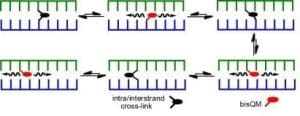
Tandem formation of QM intermediates (bisQM) provides a basis for highly efficient cross-linking of DNA since their reversible reaction offers an escape from kinetic traps that would otherwise suppress cross-linking. Even after cross-links are established, they still remain dynamic and allow for strand exchange and cross-link migration. The parameters facilitating this migration and their implications for DNA damage and repair are now under investigation.
Teaching:
030.316 Biochemistry II
030.619 Chemical Biology I
030.620 Chemical Biology II
030.613 CBI Program Forum I
030.614 CBI Program Forum II
Selected publications
A more complete list of publications can be found at http://www.researcherid.com/rid/C-4793-2009
Jimin Hu, Qi Su, Jamie Schlessman and Steven E. Rokita “Redox Control of Iodotyrosine Deiodinase.” Protein Science 2019, 28, 68-78. (https://doi.org/10.1002/pro.3479)
Zuodong Sun and Steven E. Rokita “Towards a Halophenol Dehalogenase from Iodotyrosine Deiodinase via Computational Design.” ACS Catalysis 2018, 8, 11783–11793 (DOI: 10.1021/acscatal.8b03587)
Abhishek Phatarphekar, Qi Su, Suk Ho Eun, Xin Chen, and Steven E. Rokita “The Importance of a Halotyrosine Dehalogenase for Drosophila Fertility.” J. Biol. Chem. 2018, 293, 10314–10321. (DOI 10.1074/jbc.RA118.003364)
Zuodong Sun, Qi Su, Steven E Rokita “The Distribution and Mechanism of Iodotyrosine Deiodinase Defied Expectations” Arch. Biochem. Biophys. 2017, 632, 77-87. (DOI: 10.1016/j.abb.2017.07.019).
Qi Su, Petrina A. Boucher, and Steven E. Rokita “Conversion of a Dehalogenase to a Nitroreductase by Swapping its Flavin Cofactor with a 5-Deazaflavin Analog.” Angew. Chem. Int. Ed. 2017, 56, 10862-10866 (DOI: 10.1002/anie.201703628).
Nattha Ingavat, Jennifer M. Kavran, Zuodong Sun, and Steven E. Rokita* “Active Site Binding is not Sufficient for Reductive Deiodination by Iodotyrosine Deiodinase” Biochemistry 2017, 56, 1130-1139. (DOI: 10.1021/acs.biochem.6b01308)
Abhishek Phatarphekar, and Steven E. Rokita "Functional Analysis of Iodotyrosine Deiodinase From Drosophila melanogaster" Protein Science 2016, 25, 2187-2195. (http://onlinelibrary.wiley.com/doi/10.1002/pro.3044/full)
Shalini Saha and Steven E. Rokita “An activator of an Adenylation Domain Revealed by Activity but not Sequence Homology” ChemBioChem 2016, 17, 1818-1823. (http://dx.doi.org/10.1002/cbic.201600291)
Chengyun Huang, Yang Liu and Steven E. Rokita “Targeting Duplex DNA with the Reversible Reactivity of Quinone Methides” Signal Transduct Target Ther. 2016, 1, 16009. (doi:10.1038/sigtrans.2016.9, http://www.nature.com/articles/sigtrans20169)
Arnab Mukherjee and Steven Rokita “A Single Amino Acid Switch Between a Flavin- Dependent Dehalogenase and Nitroreductase” J. Am. Chem. Soc. 2015, 137, 15342–15345. [hyperlink to: http://pubs.acs.org/doi/10.1021/jacs.5b07540]
Kostyantyn Bobyk, David Ballou and Steven Rokita “Pre-Steady-State Characterization of Iodotyrosine Deiodiase from Human Thyroid” Biochemistry 2015, 54, 4487–4494. [hyperlink to http://pubs.acs.org/doi/abs/10.1021/acs.biochem.5b00410]
Jimin Hu, Watchalee Chuenchor, and Steven Rokita “A Switch Between One- and Two-Electron Chemistry of the Human Flavoprotein Iodotyrosine Deiodinase is Controlled by Substrate” J. Biol. Chem. 2015, 290, 590-600. [hyperlink to: http://www.jbc.org/content/290/1/590.long]
Fazel Fakhari and Steven Rokita “A Walk Along DNA Using Bipedal Migration of a Dynamic and Covalent Cross-linker” Nature Commun. 2014, 5, 5591. [hyperlink to: http://www.nature.com/ncomms/2014/141121/ncomms6591/full/ncomms6591.html]
Michael McCrane Mark Hutchinson, Omer Ad, and Steven E. Rokita “Oxidative Quenching of Quinone Methide Adducts Reveals Transient Products of Reversible Alkylation in Duplex DNA” Chem. Res. Toxicol. 2014, 27, 1282–1293. [hyperlink to: http://pubs.acs.org/doi/abs/10.1021/tx500152d]
Abhishek Phatarphekar, Jennifer Buss, and Steven Rokita "Iodotyrosine Deiodinase: A Unique Flavoprotein Present in Organisms of Diverse Phyla” Mol. BioSyst. 2014,10, 86-92. [hyperlink to: http://xlink.rsc.org/?doi=C3MB70398C]
Fazel Fakhari, Yun-Yun K. Chen and Steven Rokita “Enhancing Excess Electron Transport in DNA” Chem. Commun. 2013, 49, 7073-7075. [hyperlink to http://xlink.rsc.org/?doi=C3CC43887B]
Amethist Finch, William Davis and Steven Rokita “Accumulation of the Cyclobutane Thymine Dimer in Defined Sequences of Free and Nucleosomal DNA” Photochem. Photobiol. Sci. 2013, 12, 1474–1482. [hyperlink to http://xlink.rsc.org/?doi=C3PP50147G]
Yang Liu and Steven Rokita “Inducible Alkylation of DNA by a Quinone Methide-Peptide Nucleic Acid Conjugate” Biochemistry 2012, 51, 1020-1027. [hyperlink to: http://pubs.acs.org/doi/abs/10.1021/bi201492b]